{{
}}
Faire une présentation scientifique
Joseph Salmon
- Sources d’inspiration pour cette présentation:
Quelques exemples de présentations scientifiques:
- Gabriel Peyré: excellentes diapositives ! (mathématiques appliquées)
- Persi Diaconis (Stanford Univ.) : excellent orateur (randomness)
- Sebastian Wild: sorting excellentes diapositive (algorithmes de tri)
- David Donoho : excellent orateur (statistiques)
- Emmanuel Candès: excellent orateur (statistiques)
- Hannah Fry: excellente oratrice (mathématiques appliquées)
Quatre étapes pour un bon exposé
Étape 1:
Choisir un fil narratif
Étape 2:
Planifiez vos diapositives
Étape 3:
Création des diapositives
Étape 4:
Répéter et ajuster
Étape 1: Choisir un fil narratif
Créez un plan : ne pas oublier de détails importants
Introduction (difficile): perspective large / prépare l’audience à ce qui suivra
- Utiliser une image/video pour capter l’attention
Méthodes : Décrire les méthodes utilisées (crédibilité)
- Utiliser des visuels pour décrire les méthodes
- Inclure assez de détails pour que l’audience comprenne (éviter le superflu)
Résultats : Présenter les preuves soutenant votre travail
- Choisir les résultats les plus importants et intéressants
- Décomposer les résultats complexes en parties digestes
Conclusion & Perspectives
Étape 2: Planifiez vos diapositives
Titres des diapositives : Une idée principale par diapositive
Conception des diapositives :
- Choisir le type: Quarto, Beamer, Impress (éviter PowerPoint, keynote)
- Décider comment transmettre l’idée principale : Figures, photos, équations, statistiques, références, etc.
- Corps de la diapositive : Soutenir l’idée principale
Contenu des diapositives : Lister les points à aborder en bullet points
Étape 3: Création des diapositives
- Format des diapositives :
- Diapositive de titre : Informative, inclure noms et une image attrayante
- Mise en page : Simple, beaucoup d’espace vide, ne pas surcharger
- Éléments textuels :
- Texte concis : Éviter phrases complètes, pas plus de 10-12 lignes / diapositive
- Relecture : orthographe / grammaire
- Citations et crédit: Inclure les sources
- Graphiques /Figures (mieux qu’un tableau)
- Adapter pour la présentation, éviter détails trop petits
- Couleurs : Utiliser de manière significative (attention aux daltoniens!)
- Inkscape : Pour les figures / croquis
- Transitions & animations : transitions entre sections, éviter les distractions
Étape 4: Répéter et ajuster
- Première répétition :
- Seul : Vérifiez la fluidité de votre narration
- Points à vérifier : Début et fin, transitions, animations, aide des diapositives
- Pratique devant un public :
- Devant des pairs : Chronométrer le discours
- Feedback : Noter les retours et les questions posées
- Intégrer le feedback : Supprimer les points superflues
- Répéter plusieurs fois :
- Mémoriser l’ordre : Connaître les transitions clés
- Ne pas apprendre par cœur : sauf l’ouverture, la clôture et les points clés
- Améliorer la présentation :
- Intonation et vitesse : Faire des pauses, regarder l’audience imaginaire
- Respecter le temps imparti : Laisser du temps pour les questions
- Anticiper les questions : Préparer des diapositives supplémentaires
Conseils: mise en page (10 points sur 20)
- Taille de police : Ajuster pour que 10 mots tiennent horizontalement en gros
- Polices : Ne pas utiliser plus de 2 ou 3 polices différentes
- Texte essentiel : Supprimer tout texte superflu
- Acronymes : Rappeler leur signification à chaque fois
- Majuscules : Éviter les titres en majuscules.
- Bannières et logos : Éviter les bannières, logos ou arrière-plans répétés
- Transitions : Éviter les transitions avec effets de fondu, texte rebondissant ou sons (distraction)
- Couleurs : Éviter de mélanger vert et rouge pour les figures (daltonisme).
- Figures >> tables : Utiliser des figures plutôt que des tableaux
- Légendes : Supprimer les légendes automatiques, étiqueter directement le graphique
- Lisibilité : Assurer que les polices et les largeurs de ligne sont lisibles après redimensionnement
- Données 3D : Ne jamais afficher des données 2D en 3D!
Conseils: présentation (10 points sur 20)
- Connexion : connecter votre ordinateur au projecteur (prenez votre propre connecteur si besoin)
- Notes : Éviter de lire, de faire des phrases et blagues pré-écrites
- Pointeur : Utiliser un pointeur ou votre doigt pour orienter l’audience vers des parties spécifiques de la diapositive
- Pointeur sur l’écran : Toucher l’image sur l’écran plutôt que de projeter une ombre avec le pointeur.
- Dynamisme : Parler plus fort et devenir plus dynamique si l’audience commence à fermer les yeux
- Questions : Reformuler les questions à haute voix pour le bénéfice de tous
- Mots parasites : Minimiser l’utilisation de “OK”, “euh”, “ahhh”, “er”, etc.
- Humour : Éviter le plus possible, rester professionnel
Real talk part
Mainly joint work with:
- Tanguy Lefort (Univ. Montpellier, IMAG)
- Benjamin Charlier (Univ. Montpellier, IMAG)
- Camille Garcin (Univ. Montpellier, IMAG)
- Maximilien Servajean (Univ. Paul-Valéry-Montpellier, LIRMM, Univ. Montpellier)
- Alexis Joly (Inria, LIRMM, Univ. Montpellier)
and from
- Pierre Bonnet, Hervé Goëau (CIRAD, AMAP)
- Antoine Affouard, Jean-Christophe Lombardo, Titouan Lorieul, Mathias Chouet (Inria, LIRMM, Univ. Montpellier)
Pl@ntNet description
Pl@ntNet: ML for citizen science
A citizen science platform using machine learning to help people identify plants with their mobile phones

- Website: https://plantnet.org/
- Note: no mushroom identification!
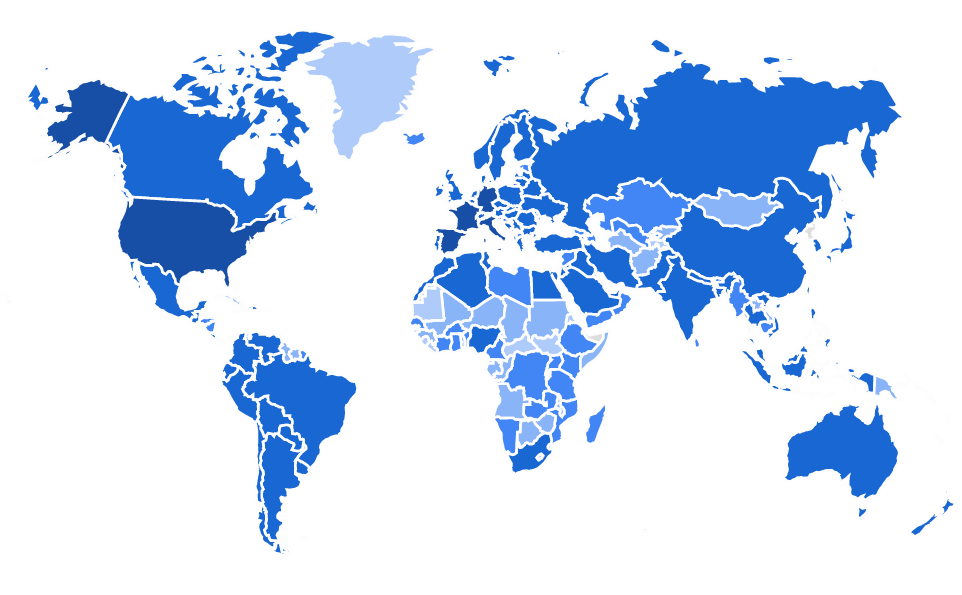
https://identify.plantnet.org/stats
- Start in 2011, now 25M+ users
- 200+ countries
- Up to 2M image uploaded/day
- 50K species
- 1B+ total images
- 10M+ labeled / validated
Pl@ntNet & Cooperative Learning
Note: I am mostly innocent; started working with the Pl@ntNet team in 2020
Motivation: excellent app … but not a perfect app; How to improve?
- Community effort: machine learning, ecology, engineering, amateurs
- Many open problems (theoretical/practical)
- Need for methodological/computational breakthrough
Contributions
- Pl@ntNet-300K (Garcin et al. 2021): Creation and release of a large-scale dataset sharing the same property as Pl@ntNet; available for the community to improve learning systems
- Learning & crowd-sourced data (Lefort et al. 2024) and (Lefort et al. 2025): How to leverage multiple labels per image to improve the model? Need to assert quality: the workers, the images/labels, the model, etc.
- Top-K learning (Garcin et al. 2022): Driven by theory, introduce new loss to cope with Pl@ntNet constraints to output multiple labels (e.g., UX, Deep Learning framework, etc.)
Dataset release: Pl@ntNet-300K
Popular datasets limitations:
- structure of labels too simplistic (CIFAR-10, CIFAR-100)
- might have tasks too easy to discriminate
- might be too well-balanced (same number of images per class)
- contains duplicate, low-quality, or irrelevant images
Motivation:
release a large-scale dataset sharing similar features as the Pl@ntNet dataset to foster research in plant identification
\(\implies\) Pl@ntNet-300K (Garcin et al. 2021)
Intra-class variability










Inter-class ambiguity










Sampling bias
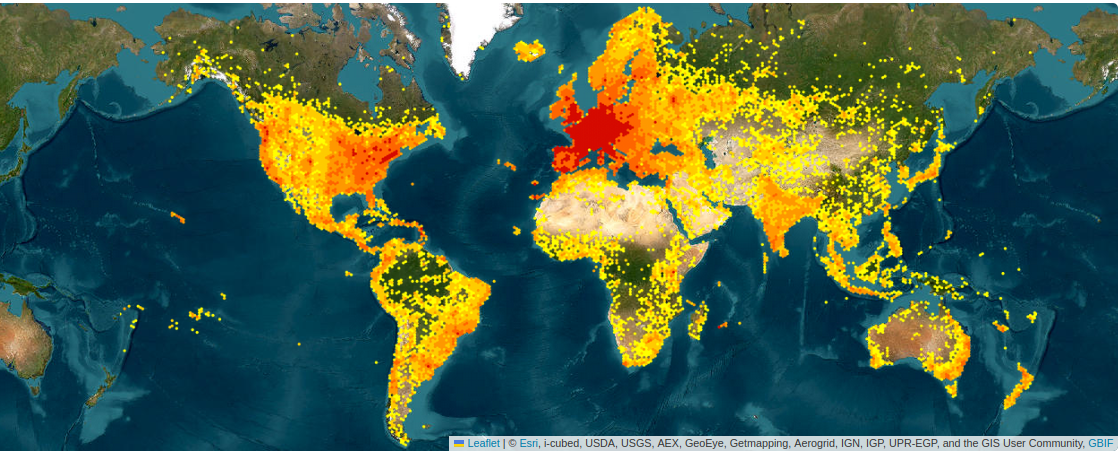
Top-5 most observed plant species in Pl@ntNet (13/04/2024):
25134 obs.  Echium vulgare L.
Echium vulgare L.
24720 obs.  Ranunculus ficaria L.
Ranunculus ficaria L.
24103 obs.  Prunus spinosa L.
Prunus spinosa L.
23288 obs.  Zea mays L.
Zea mays L.
23075 obs.  Alliaria petiolata
Alliaria petiolata
10753 obs.

Centaurea jacea
6 obs.

Cenchrus agrimonioides
8376 obs.

Magnolia grandiflora
413 obs.

Moehringia trinervia
Many more biases …
- Selection bias
- Convenience sampling: easily vs. hardly accessible
- Preference for certain species: visibility / ease of identification
- Subjective bias: selection based on personal judgment, may not be random or representative
- Rare species: rare or endangered species may be under-represented
- Temporal bias / seasonal variation: seasonal changes in plant characteristics
- …
Construction of Pl@ntNet-300K
- Earth: 300K+ species
- Pl@ntNet: 50K+ species
- Pl@ntNet-300K: 1K+ species
Note: long tail preserved by genera subsampling
Caracteristics:
- 306,146 color images
- Size : 32 GB
- Labels: K=1,081 species
- Required 2,079,003 volunteers “workers”
Zenodo, 1 click download
https://zenodo.org/record/5645731
Code to train models
https://github.com/plantnet/PlantNet-300KVotes, labels & aggregation
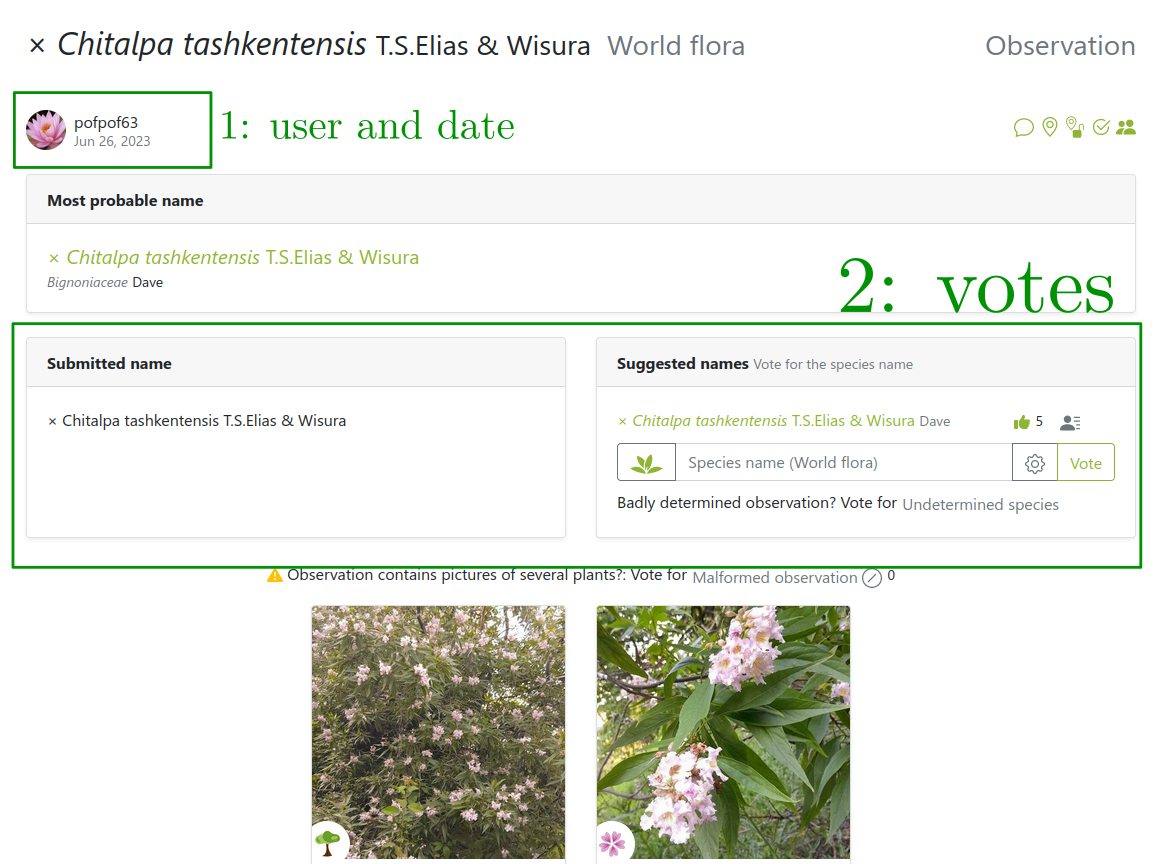
Images from users… so are the labels!
But users can be wrong or not experts
Several labels can be available per image!
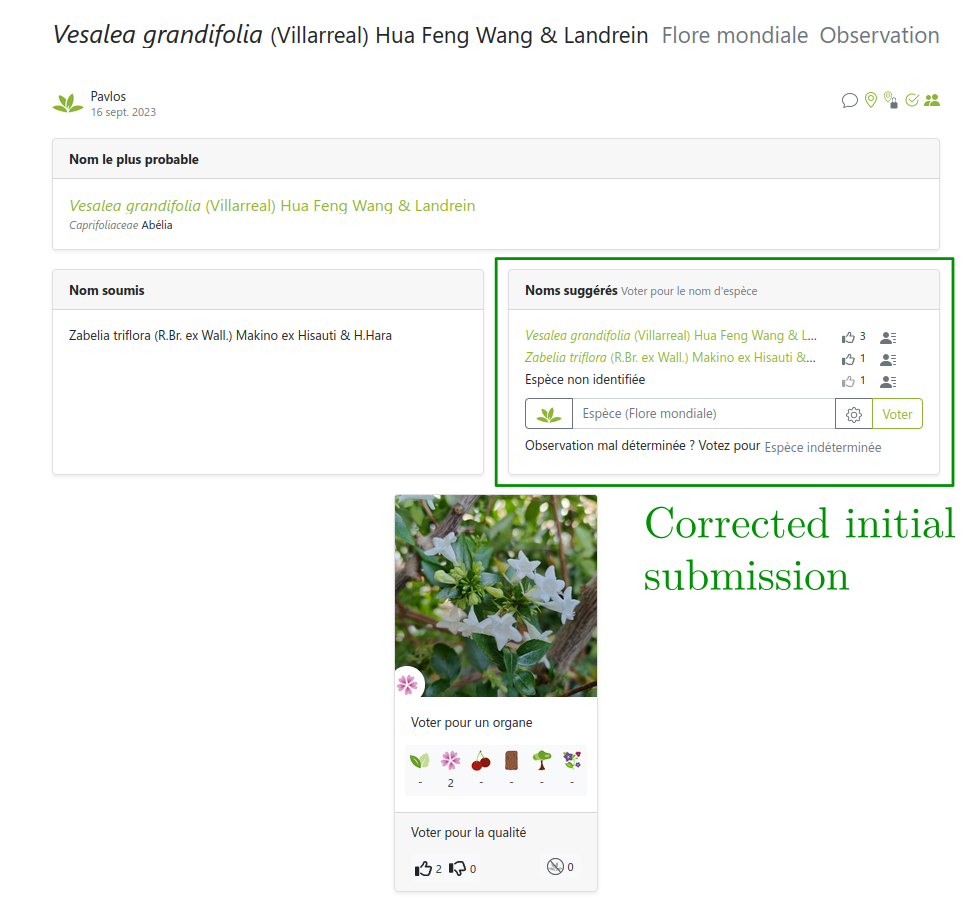
But sometimes users can’t be trusted
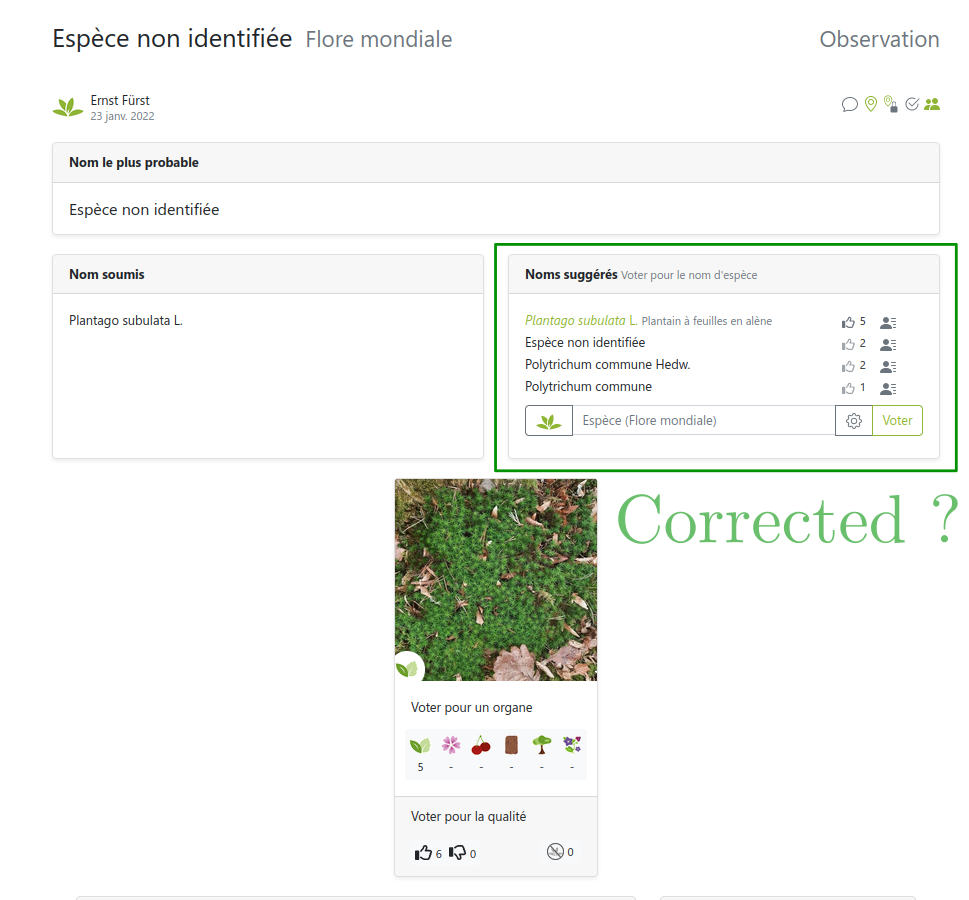
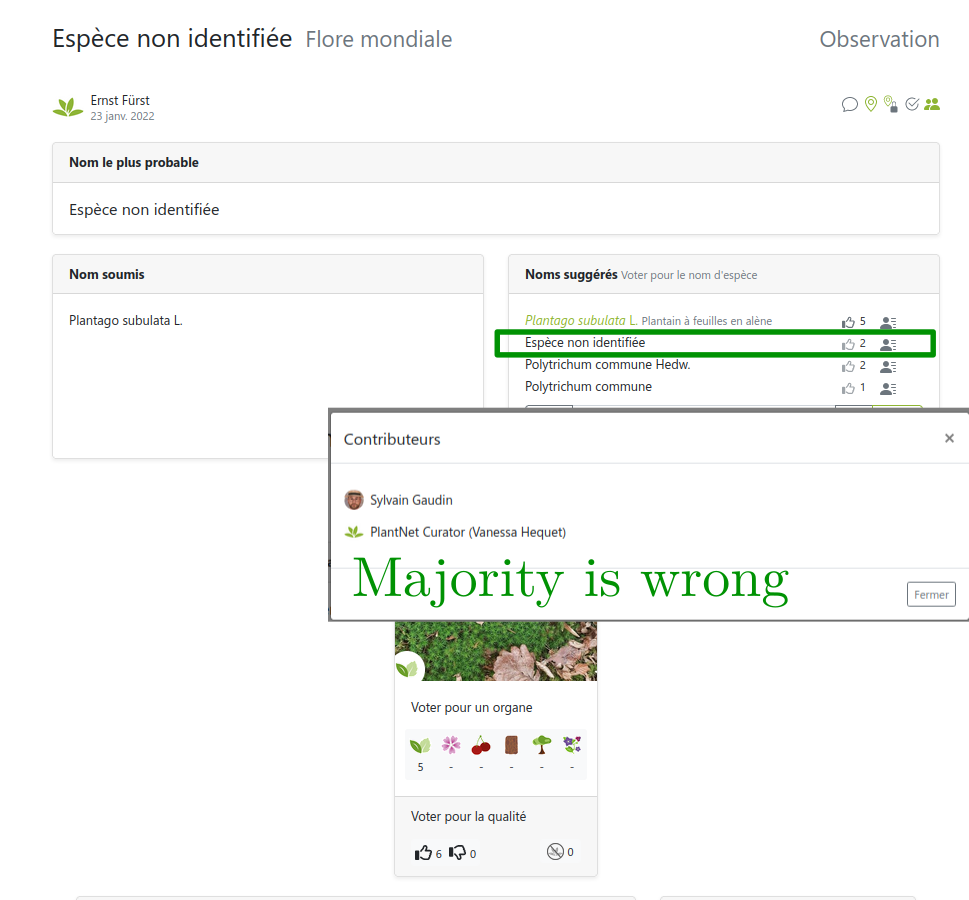
Link: https://identify.plantnet.org/weurope/observations/1012500059
The good, the bad and the ugly
- The good: fast, easy, cheap data collection
- The bad: noisy labels with different levels of expertise
- The ugly: (partly) missing theory, ad-hoc methods for noisy labels
(Weighted) Majority Vote
Objective
Provide for all images \(x_i\) an aggregated label \(\hat{y}_i\) based on the votes \(y^{u}_i\) of the workers \(u \in \mathcal{U}\).
Majority Vote (MV): intuitively
Naive idea: make users vote and take the most voted label for each image
Majority Vote : formally
Naive idea: make users vote and take the most voted label for each image
\[ \forall x_i \in \mathcal{X}_{\text{train}},\quad \hat y_i^{\text{MV}} = \mathop{\mathrm{arg\,max}}_{k\in [K]} \Big(\sum\limits_{u\in\mathcal{U}(x_i)} {1\hspace{-3.8pt} 1}_{\{y^{u}_i=k\}} \Big) \]
Properties:
✓ simple
✓ adapted for any number of users
✓ efficient, few labelers sufficient (say < 5, Snow et al. 2008)
✗ ineffective for borderline cases
✗ suffer from spammers / adversarial users
Constraints: wide range of skills, different levels of expertise
Modeling aspect: add a user weight to balance votes
Assume given weights \((w_u)_{u\in\mathcal{U}}\) for now
Weighted Majority Vote (WMV): example
The label confidence \(\mathrm{conf}_{i}(k)\) of label \(k\) for image \(x_i\) is the sum of the weights of the workers who voted for \(k\): \[ \forall k \in [K], \quad \mathrm{conf}_{i}(k) = \sum\limits_{u\in\mathcal{U}(x_i)} w_u {1\hspace{-3.8pt} 1}_{\{y^{u}_i=k\}} \]
Size effect:
- more votes \(\Rightarrow\) more confidence
- more expertise \(\Rightarrow\) more confidence
The label accuracy \(\mathrm{acc}_{i}(k)\) of label \(k\) for image \(x_i\) is the normalized sum of weights of the workers who voted for \(k\): \[ \forall k \in [K], \quad \mathrm{acc}_{i}(k) = \frac{\mathrm{conf}_i(k)}{\sum\limits_{k'\in [K]} \mathrm{conf}_i(k')} \]
Interpretation:
- only the proportion of the weights matters
Weighted Majority Vote (WMV)
Majority voting but weighted by a confidence score per user \(u\): \[ \forall x_i \in \mathcal{X}_{\texttt{train}},\quad \hat{y}_i^{\textrm{WMV}} = \mathop{\mathrm{arg\,max}}_{k\in [K]} \Big(\sum\limits_{u\in\mathcal{U}(x_i)} w_u {1\hspace{-3.8pt} 1}_{\{y^{u}_i=k\}} \Big) \]
Note: the weighted majority vote can be computed from confidence or accuracy \[ \hat{y}_i^{\textrm{WMV}} = \mathop{\mathrm{arg\,max}}_{k\in [K]} \Big( \mathrm{conf}_i(k) \Big) = \mathop{\mathrm{arg\,max}}_{k\in [K]} \Big(\mathrm{acc}_i(k) \Big) \]
Two pillars for validating a label \(\hat{y}_i\) for an image \(x_i\) in Pl@ntNet :
Expertise: labels quality check
keep images with label confidence above a threshold \(\theta_{\text{conf}}\), validate \(\hat{y}_i\) when \[ \boxed{\mathrm{conf}_{i}(\hat{y}_i) > \theta_{\text{conf}}} \]
Consensus: labels agreement check
keep images with label accuracy above a threshold \(\theta_{\text{acc}}\), validate \(\hat{y}_i\) when \[ \boxed{\mathrm{acc}_{i}(\hat{y}_i) > \theta_{\text{acc}}} \]
Pl@ntNet label aggregation
(EM algorithm)
Weighting scheme: weight user vote by its number of identified species
Weights example
- \(n_{\mathrm{user}} = 6\)
- \(K=3\) : Rosa indica, Ficus elastica, Mentha arvensis
- \(\theta_{\text{conf}}=2\) and \(\theta_{\text{acc}}=0.7\)
Take into account 4 users out of 6
Take into account 4 users out of 6
Invalidated label: Adding User 5 reduces accuracy
Label switched: User 6 is an expert (even self-validating)
Choice of weight function
\[ f(n_u) = n_u^\alpha - n_u^\beta + \gamma \text{ with } \begin{cases} \alpha = 0.5 \\ \beta=0.2 \\ \gamma=\log(1.7)\simeq 0.74 \end{cases} \]
Other existing strategies
- Majority Vote (MV)
Worker agreement with aggregate (WAWA): 2-step method
- Majority vote
- Weight users by how much they agree with the majority
- Weighted majority vote
- TwoThrid (iNaturalist)
- Need 2 votes
- \(2/3\) of agreements
Pl@ntNet labels release: South West. European flora
Extracting a subset of a Pl@ntNet votes
- South Western European flora observations since \(2017\)
- \(~800K\) users answered more than \(11K\) species
- \(~6.6M\) observations
- \(~9M\) votes casted
- Imbalance: 80% of observations are represented by 10% of total votes
No ground truth available to evaluate the strategies
Test sets without ground truth
- Extract \(98\) experts: Tela Botanica + prior knowledge (P. Bonnet)
Pl@ntNet South Western European flora
Accuracy and number of classes kept
- Pl@ntNet aggregation performs better overall
- TwoThird is highly impacted by the reject threshold
- In ambiguous settings, strategies weighting users are better
Performance: Precision, recall and validity
- Pl@ntNet aggregation performs better overall
- TwoThird has good precision but bad recall
- We indeed remove some data but less than TwoThird
Why?
- More data
- Could correct non-expert users
- Could invalidate bad quality observation
Main danger
- Model collapse (Shumailov et al. 2024): users are already guided by AI predictions
Potential strategies to integrate the AI vote
- AI as worker: naive integration
- AI fixed weight:
- weight fixed to \(1.7\)
- can invalidate two new users but is not self-validating
- AI invalidating:
- weight fixed to \(1.7\)
- can only invalidate observation
- AI confident:
- weight fixed to \(1.7\)
- can participate if confidence in prediction high enough (\(\theta_{\text{score}}\))
\(\Longrightarrow\) confident AI with \(\theta_{\text{score}}=0.7\) performs best… but invalidating AI could be preferred for safety \(\Longleftarrow\)
Aggregating labels: open source tool
peerannot: Python library to handle crowdsourced data
Conclusion
- Challenges in citizen science: many and varied (need more attention)
- Crowdsourcing / Label uncertainty: helpful for data curation
- Improved data quality \(\implies\) improved learning performance
Dataset release:
- Pl@ntNet-300K: https://zenodo.org/record/5645731
- Pl@ntNet SWE flora: https://zenodo.org/records/10782465
Code release:
- Toolbox: https://peerannot.github.io/
- Some benchmarks: https://benchopt.github.io/
Future work
- Uncertainty quantification
- Improve robustness to adversarial users
- Leverage gamification for more quality labels theplantgame.com

