%matplotlib inline
import os
import numpy as np
import calendar
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from cycler import cycler
import pooch # download data / avoid re-downloading
from IPython import get_ipython
sns.set_palette("colorblind")
palette = sns.color_palette("twilight", n_colors=12)
pd.options.display.max_rows = 8Disclaimer: this course is adapted from the work Pandas tutorial by Joris Van den Bossche. R users might also want to read Pandas: Comparison with R / R libraries for a smooth start in Pandas.
We start by importing the necessary libraries:
Data preparation
Automatic data download
First, it is important to download automatically remote files for reproducibility (and avoid typing names manually). Let us apply this to the Titanic dataset:
url = "http://josephsalmon.eu/enseignement/datasets/titanic.csv"
path_target = "./titanic.csv"
path, fname = os.path.split(path_target)
pooch.retrieve(url, path=path, fname=fname, known_hash=None) # if needed `pip install pooch`'/home/jsalmon/Documents/Mes_cours/Montpellier/HAX712X/Courses/Pandas/titanic.csv'Reading the file as a pandas dataframe with read_csv:
df_titanic_raw = pd.read_csv("titanic.csv")Visualize the end of the dataset:
df_titanic_raw.tail(n=3)| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 888 | 889 | 0 | 3 | Johnston, Miss. Catherine Helen "Carrie" | female | NaN | 1 | 2 | W./C. 6607 | 23.45 | NaN | S |
| 889 | 890 | 1 | 1 | Behr, Mr. Karl Howell | male | 26.0 | 0 | 0 | 111369 | 30.00 | C148 | C |
| 890 | 891 | 0 | 3 | Dooley, Mr. Patrick | male | 32.0 | 0 | 0 | 370376 | 7.75 | NaN | Q |
Visualize the beginning of the dataset:
df_titanic_raw.head(n=5)| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
Missing values
It is common to encounter features/covariates with missing values. In pandas they were mostly handled as np.nan (not a number) in the past. With versions >=1.0 missing values are now treated as NA (Not Available), in a similar way as in R; see the Pandas documentation on missing data for standard behavior and details.
Note that the main difference between pd.NA and np.nan is that pd.NA propagates even for comparisons:
pd.NA == 1<NA>whereas
np.nan == 1FalseTesting the presence of missing values
print(pd.isna(pd.NA), pd.isna(np.nan))True TrueThe simplest strategy (when you can / when you have enough samples) consists of removing all nans/NAs, and can be performed with dropna:
df_titanic = df_titanic_raw.dropna()
df_titanic.tail(3)| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 879 | 880 | 1 | 1 | Potter, Mrs. Thomas Jr (Lily Alexenia Wilson) | female | 56.0 | 0 | 1 | 11767 | 83.1583 | C50 | C |
| 887 | 888 | 1 | 1 | Graham, Miss. Margaret Edith | female | 19.0 | 0 | 0 | 112053 | 30.0000 | B42 | S |
| 889 | 890 | 1 | 1 | Behr, Mr. Karl Howell | male | 26.0 | 0 | 0 | 111369 | 30.0000 | C148 | C |
You can see for instance the passenger with PassengerId 888 has been removed by the dropna call.
# Useful info on the dataset (especially missing values!)
df_titanic.info()<class 'pandas.core.frame.DataFrame'>
Index: 183 entries, 1 to 889
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 183 non-null int64
1 Survived 183 non-null int64
2 Pclass 183 non-null int64
3 Name 183 non-null object
4 Sex 183 non-null object
5 Age 183 non-null float64
6 SibSp 183 non-null int64
7 Parch 183 non-null int64
8 Ticket 183 non-null object
9 Fare 183 non-null float64
10 Cabin 183 non-null object
11 Embarked 183 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 18.6+ KB# Check that the `Cabin` information is mostly missing; the same holds for `Age`
df_titanic_raw.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KBOther strategies are possible to handle missing values but they are out of scope for this preliminary course on pandas, see the documentation for more details.
Data description
Details of the dataset are given here
Survived: Survival 0 = No, 1 = YesPclass: Ticket class 1 = 1st, 2 = 2nd, 3 = 3rdSex: Sex male/femaleAge: Age in yearsSibsp: # of siblings/spouses aboard the TitanicParch: # of parents/children aboard the TitanicTicket: Ticket numberFare: Passenger fareCabin: Cabin numberEmbarked: Port of Embarkation C = Cherbourg, Q = Queenstown, S = SouthamptonName: Name of the passengerPassengerId: Number to identify passenger
For those interested, an extended version of the dataset is available here https://biostat.app.vumc.org/wiki/Main/DataSets.
Simple descriptive statistics can be obtained using the describe method:
df_titanic.describe()| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| count | 183.000000 | 183.000000 | 183.000000 | 183.000000 | 183.000000 | 183.000000 | 183.000000 |
| mean | 455.366120 | 0.672131 | 1.191257 | 35.674426 | 0.464481 | 0.475410 | 78.682469 |
| std | 247.052476 | 0.470725 | 0.515187 | 15.643866 | 0.644159 | 0.754617 | 76.347843 |
| min | 2.000000 | 0.000000 | 1.000000 | 0.920000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 263.500000 | 0.000000 | 1.000000 | 24.000000 | 0.000000 | 0.000000 | 29.700000 |
| 50% | 457.000000 | 1.000000 | 1.000000 | 36.000000 | 0.000000 | 0.000000 | 57.000000 |
| 75% | 676.000000 | 1.000000 | 1.000000 | 47.500000 | 1.000000 | 1.000000 | 90.000000 |
| max | 890.000000 | 1.000000 | 3.000000 | 80.000000 | 3.000000 | 4.000000 | 512.329200 |
Visualization
- Histograms: usually has one parameter controlling the number of bins (please avoid… often useless, when many samples are available)
fig, ax = plt.subplots(1, 1, figsize=(5, 3))
ax.hist(df_titanic['Age'], density=True, bins=25)
plt.xlabel('Age')
plt.ylabel('Proportion')
plt.title("Passager age histogram")
plt.show()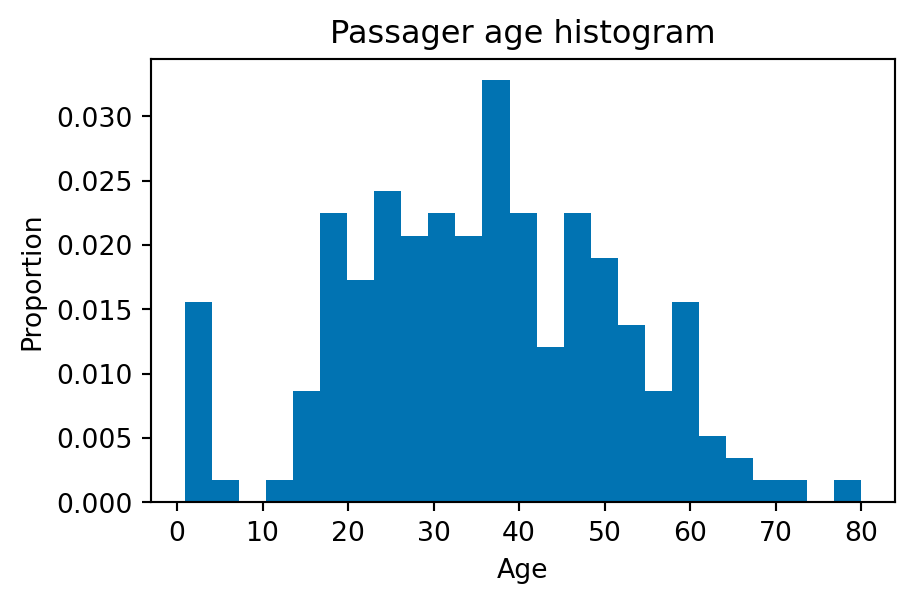
- Kernel Density Estimate (KDE): usually has one parameter (the bandwidth) controlling the smoothing level.
fig, ax = plt.subplots(1, 1, figsize=(5, 3))
sns.kdeplot(
df_titanic["Age"], ax=ax, fill=True, cut=0, bw_adjust=0.1
)
plt.xlabel("Proportion")
plt.ylabel("Age")
plt.title("Passager age kernel density estimate")
plt.tight_layout()
plt.show()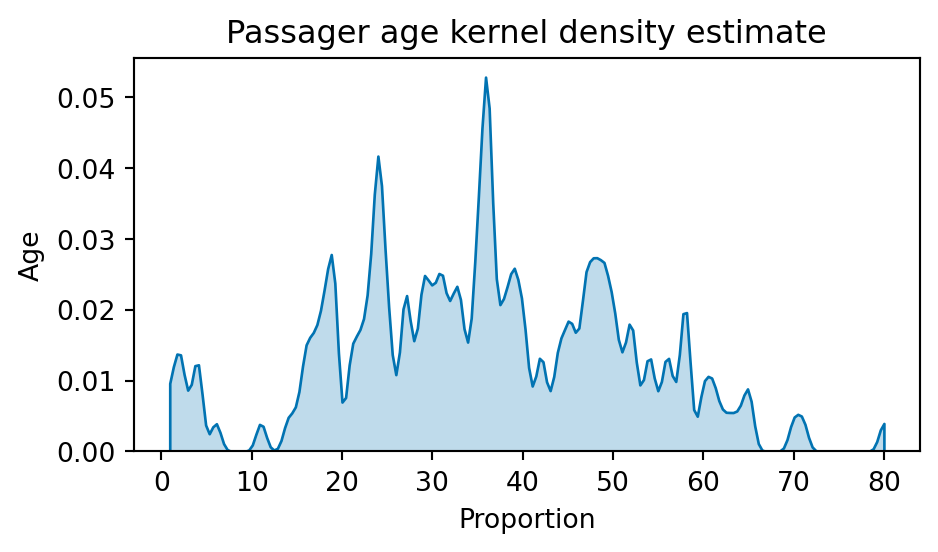
The bandwidth parameter (here encoded as bw_adjust) controls the smoothing level. It is a common parameter in kernel smoothing, investigated thoroughly in the non-parametric statistics literature.
- Swarmplot:
fig, ax = plt.subplots(1, 1, figsize=(6, 5))
sns.swarmplot(
data=df_titanic_raw,
ax=ax,
x="Sex",
y="Age",
hue="Survived",
palette={0: "r", 1: "k"},
order=["female", "male"],
)
plt.title("Passager age by gender/survival")
plt.legend(labels=["Survived", "Died"], loc="upper left")
plt.tight_layout()
plt.show()
Interactivity
Interactive interaction with codes and output is nowadays getting easier and easier (see also Shiny app in R-software). In Python, one can use plotly, or widgets and the interact package for this purpose. We are going to visualize that on the simple KDE and histogram examples.
import plotly.graph_objects as go
import numpy as np
from scipy import stats
# Create figure
fig = go.FigureWidget()
# Add traces, one for each slider step
bws = np.arange(0.01, 0.51, 0.01)
xx = np.arange(0, 100, 0.5)
for step in bws:
kernel = stats.gaussian_kde(df_titanic["Age"], bw_method=step)
yy = kernel(xx)
fig.add_trace(
go.Scatter(
visible=False,
fill="tozeroy",
fillcolor="rgba(67, 67, 67, 0.5)",
line=dict(color="black", width=2),
name=f"Bw = {step:.2f}",
x=xx,
y=yy,
)
)
# Make 10th trace visible
fig.data[10].visible = True
# Create and add slider
steps = []
for i in range(len(fig.data)):
step = dict(
method="update",
args=[{"visible": [False] * len(fig.data)}],
label=f"{bws[i]:.2f}",
)
step["args"][0]["visible"][i] = True # Toggle i'th trace to "visible"
steps.append(step)
sliders = [
dict(
active=10,
currentvalue={"prefix": "Bandwidth = "},
pad={"t": 50},
steps=steps,
)
]
fig.update_layout(
sliders=sliders,
template="simple_white",
title=f"Impact of the bandwidth on the KDE",
showlegend=False,
xaxis_title="Age"
)
fig.show()import plotly.graph_objects as go
import numpy as np
from scipy import stats
# Create figure
fig = go.FigureWidget()
# Add traces, one for each slider step
bws = np.arange(0.01, 5.51, 0.3)
xx = np.arange(0, 100, 0.5)
for step in bws:
fig.add_trace(
go.Histogram(
x=df_titanic["Age"],
xbins=dict( # bins used for histogram
start=xx.min(),
end=xx.max(),
size=step,
),
histnorm="probability density",
autobinx=False,
visible=False,
marker_color="rgba(67, 67, 67, 0.5)",
name=f"Bandwidth = {step:.2f}",
)
)
# Make 10th trace visible
fig.data[10].visible = True
# Create and add slider
steps = []
for i in range(len(fig.data)):
step = dict(
method="update",
args=[{"visible": [False] * len(fig.data)}],
label=f"{bws[i]:.2f}",
)
step["args"][0]["visible"][i] = True # Toggle i'th trace to "visible"
steps.append(step)
sliders = [
dict(
active=10,
currentvalue={"prefix": "Bandwidth = "},
pad={"t": 50},
steps=steps,
)
]
fig.update_layout(
sliders=sliders,
template="simple_white",
title=f"Impact of the bandwidth on histograms",
showlegend=False,
xaxis_title="Age"
)
fig.show()IPython widgets (in Jupyter notebooks)
import ipywidgets as widgets
from ipywidgets import interact, fixed
def hist_explore(
dataset=df_titanic,
variable=df_titanic.columns,
n_bins=24,
alpha=0.25,
density=False,
):
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
ax.hist(dataset[variable], density=density, bins=n_bins, alpha=alpha)
plt.ylabel("Density level")
plt.title(f"Dataset {dataset.attrs['name']}:\n Histogram for passengers' age")
plt.tight_layout()
plt.show()
interact(
hist_explore,
dataset=fixed(df_titanic),
n_bins=(1, 50, 1),
alpha=(0, 1, 0.1),
density=False,
)def kde_explore(dataset=df_titanic, variable=df_titanic.columns, bw=5):
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
sns.kdeplot(dataset[variable], bw_adjust=bw, fill=True, cut=0, ax=ax)
plt.ylabel("Density level")
plt.title(f"Dataset:\n KDE for passengers' {variable}")
plt.tight_layout()
plt.show()
interact(kde_explore, dataset=fixed(df_titanic), bw=(0.001, 2, 0.01))Data wrangling
groupby function
Here is an example of the using the groupby function:
df_titanic.groupby(['Sex'])[['Survived', 'Age', 'Fare', 'Pclass']].mean()| Survived | Age | Fare | Pclass | |
|---|---|---|---|---|
| Sex | ||||
| female | 0.931818 | 32.676136 | 89.000900 | 1.215909 |
| male | 0.431579 | 38.451789 | 69.124343 | 1.168421 |
You can answer other similar questions, such as “how does the survival rate change w.r.t. to sex”?
df_titanic_raw.groupby('Sex')[['Survived']].aggregate(lambda x: x.mean())| Survived | |
|---|---|
| Sex | |
| female | 0.742038 |
| male | 0.188908 |
How does the survival rate change w.r.t. the class?
df_titanic.columnsIndex(['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp',
'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked'],
dtype='object')fig, ax = plt.subplots(1, 1, figsize=(5, 5))
df_titanic.groupby('Pclass')['Survived'].aggregate(lambda x:
x.mean()).plot(ax=ax,kind='bar')
plt.xlabel('Classes')
plt.ylabel('Survival rate')
plt.title('Survival rate per classs')
plt.show()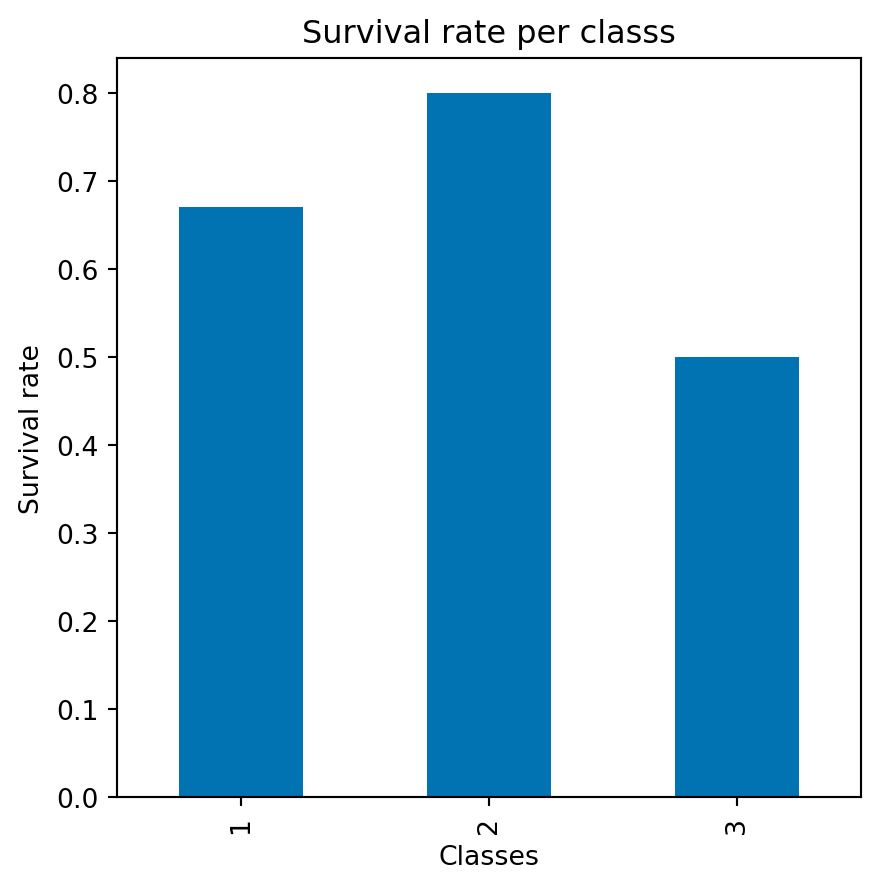
catplot: a visual groupby
ax=sns.catplot(
x="Pclass",
y="Age",
hue="Sex",
palette={'female': 'red', 'male': 'b'},
data=df_titanic_raw,
jitter = '0.2',
s=8,
)
plt.title("Sex repartition per class on the Titanic")
sns.move_legend(ax, "upper left", bbox_to_anchor=(0.8, 0.8))
plt.show()
ax=sns.catplot(
x="Pclass",
y="Age",
hue="Sex",
palette={'female': 'red', 'male': 'b'},
alpha=0.8,
data=df_titanic_raw,
kind='swarm',
s=11,
)
plt.title("Sex repartition per class on the Titanic")
sns.move_legend(ax, "upper left", bbox_to_anchor=(0.8, 0.8))
plt.show()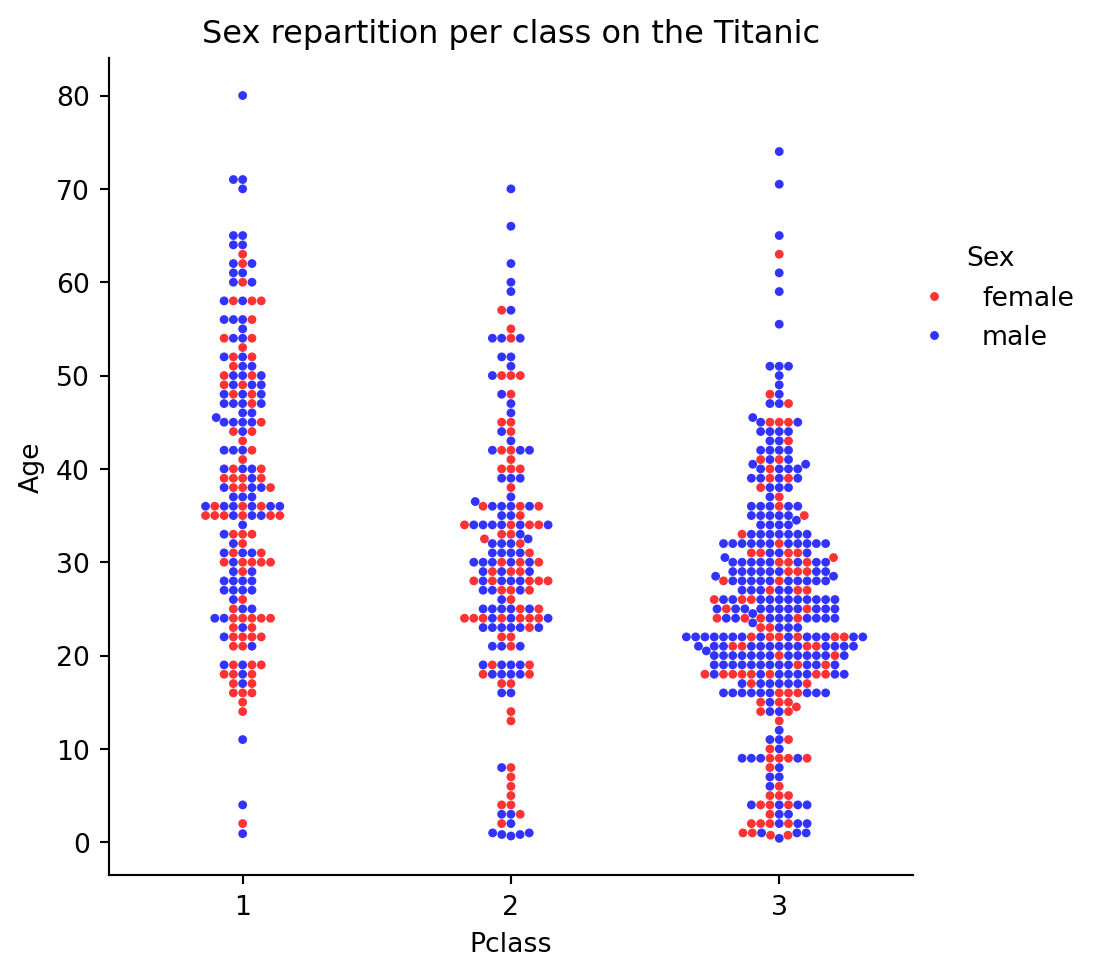
ax=sns.catplot(
x="Sex",
y="Age",
hue="Sex",
palette={'female': 'red', 'male': 'b'},
col='Pclass',
alpha=0.8,
data=df_titanic_raw,
kind='swarm',
s=6,
height=5,
aspect=0.49,
)
plt.suptitle("Sex repartition per class on Titanic")
plt.tight_layout()
plt.show()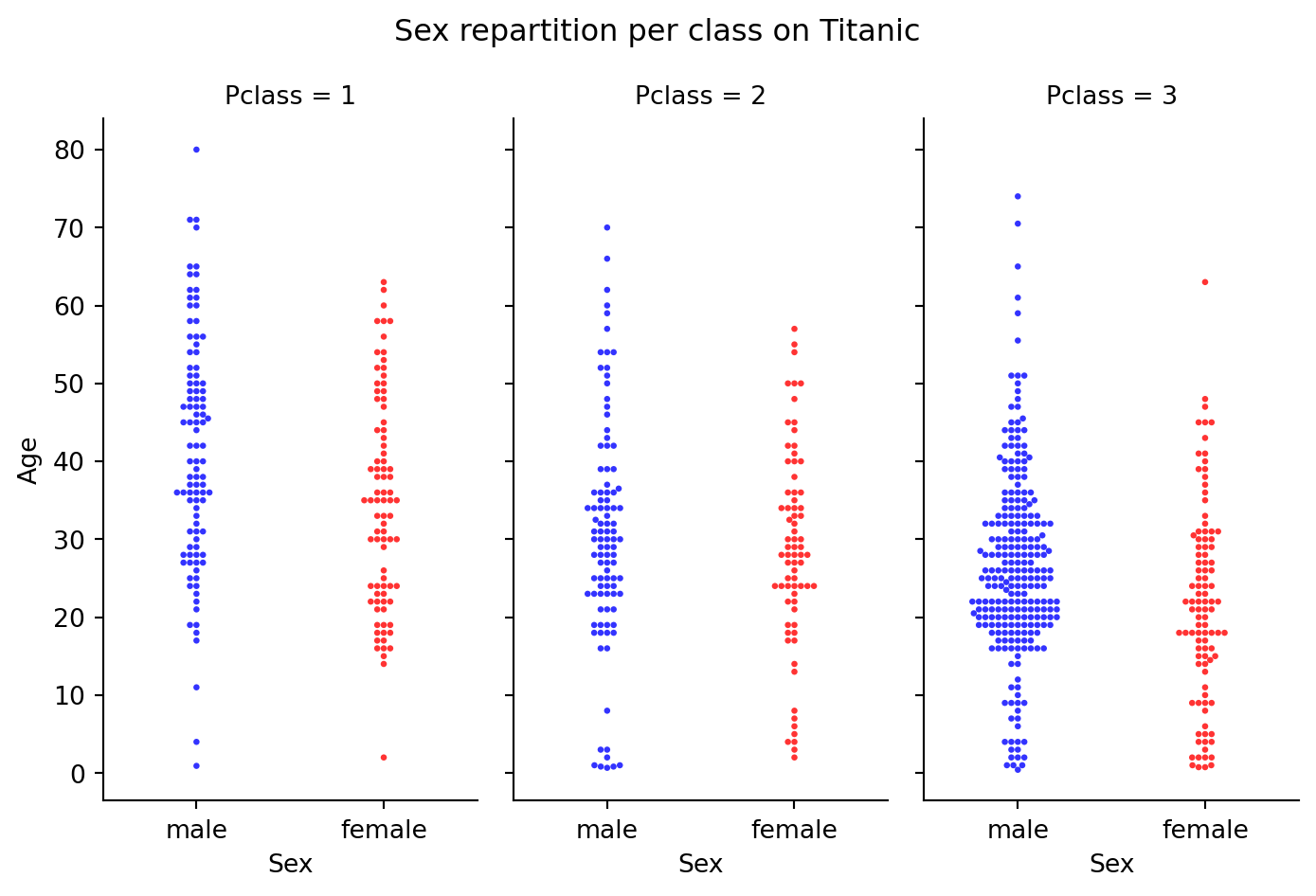
ax=sns.catplot(x='Pclass',
y='Age',
hue="Sex",
palette={'female': 'red', 'male': 'b'},
data=df_titanic_raw,
kind="violin",
alpha=0.8,
)
plt.title("Sex repartition per class on the Titanic")
sns.move_legend(ax, "upper left", bbox_to_anchor=(0.8, 0.8))
plt.show()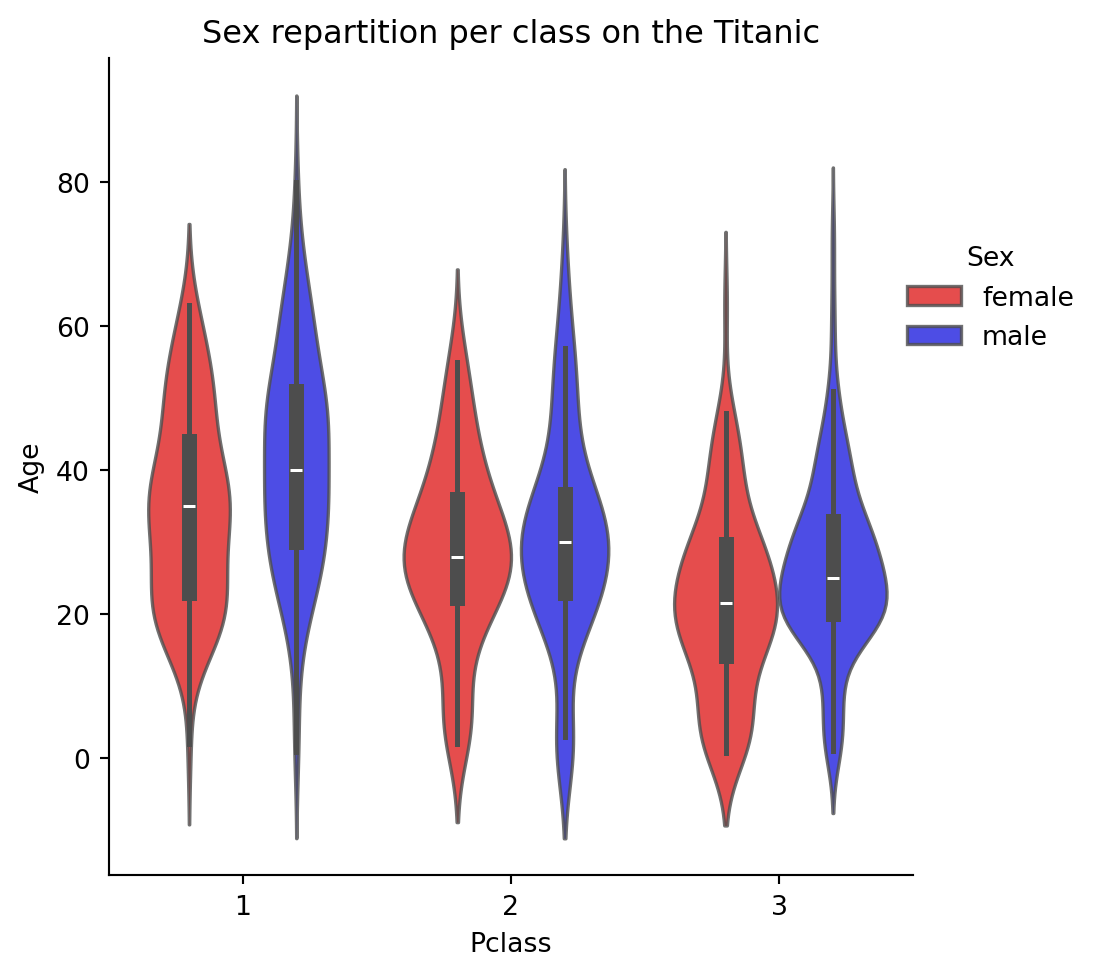
Beware: there is a large difference in sex ratio by class. You can use groups to get the counts illustrating this:
Raw sex repartition per class:
df_titanic_raw.groupby(['Sex', 'Pclass'])[['Sex']].count()| Sex | ||
|---|---|---|
| Sex | Pclass | |
| female | 1 | 94 |
| 2 | 76 | |
| 3 | 144 | |
| male | 1 | 122 |
| 2 | 108 | |
| 3 | 347 |
Raw sex ratio:
df_titanic.groupby(['Sex', 'Pclass'])[['Sex']].count()| Sex | ||
|---|---|---|
| Sex | Pclass | |
| female | 1 | 74 |
| 2 | 9 | |
| 3 | 5 | |
| male | 1 | 84 |
| 2 | 6 | |
| 3 | 5 |
Sex repartition per class (without missing values):
df_titanic_raw.groupby(['Sex'])[['Sex']].count()| Sex | |
|---|---|
| Sex | |
| female | 314 |
| male | 577 |
Raw sex ratio (without missing values):
df_titanic.groupby(['Sex'])[['Sex']].count()| Sex | |
|---|---|
| Sex | |
| female | 88 |
| male | 95 |
Consider checking the raw data, as often boxplots or simple statistics are not enough to understand the diversity inside the data; see for instance the discussion by Carl Bergstrom on Mastodon.
References:
- Practical Business Python, Comprehensive Guide to Grouping and Aggregating with Pandas, by Chris Moffitt
pd.crosstab
pd.crosstab(
df_titanic_raw["Sex"],
df_titanic_raw["Pclass"],
values=df_titanic_raw["Sex"],
aggfunc="count",
normalize=True,
)| Pclass | 1 | 2 | 3 |
|---|---|---|---|
| Sex | |||
| female | 0.105499 | 0.085297 | 0.161616 |
| male | 0.136925 | 0.121212 | 0.389450 |
Listing rows and columns
df_titanic.indexIndex([ 1, 3, 6, 10, 11, 21, 23, 27, 52, 54,
...
835, 853, 857, 862, 867, 871, 872, 879, 887, 889],
dtype='int64', length=183)df_titanic.columnsIndex(['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp',
'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked'],
dtype='object')From numpy to pandas
useful for using packages on top of pandas (e.g., sklearn, though nowadays it works out of the box with pandas)
array_titanic = df_titanic.values # associated numpy array
array_titanicarray([[2, 1, 1, ..., 71.2833, 'C85', 'C'],
[4, 1, 1, ..., 53.1, 'C123', 'S'],
[7, 0, 1, ..., 51.8625, 'E46', 'S'],
...,
[880, 1, 1, ..., 83.1583, 'C50', 'C'],
[888, 1, 1, ..., 30.0, 'B42', 'S'],
[890, 1, 1, ..., 30.0, 'C148', 'C']], dtype=object)pandas DataFrame
1D dataset: Series (a column of a DataFrame)
A Series is a labeled 1D column of a kind.
fare = df_titanic['Fare']
fare1 71.2833
3 53.1000
6 51.8625
10 16.7000
...
872 5.0000
879 83.1583
887 30.0000
889 30.0000
Name: Fare, Length: 183, dtype: float64Attributes Series: indices and values
fare.values[:10]array([ 71.2833, 53.1 , 51.8625, 16.7 , 26.55 , 13. ,
35.5 , 263. , 76.7292, 61.9792])Contrarily to numpy arrays, you can index with other formats than integers (the underlying structure being a dictionary):
head command
df_titanic_raw.head(3)| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
# Be careful, what follows changes the indexing
df_titanic_raw = df_titanic_raw.set_index('Name')
df_titanic_raw.head(3)| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | |||||||||||
| Braund, Mr. Owen Harris | 1 | 0 | 3 | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| Cumings, Mrs. John Bradley (Florence Briggs Thayer) | 2 | 1 | 1 | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| Heikkinen, Miss. Laina | 3 | 1 | 3 | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
tail command
df_titanic_raw.head(3)| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | |||||||||||
| Braund, Mr. Owen Harris | 1 | 0 | 3 | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| Cumings, Mrs. John Bradley (Florence Briggs Thayer) | 2 | 1 | 1 | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| Heikkinen, Miss. Laina | 3 | 1 | 3 | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
age = df_titanic_raw['Age']
age['Behr, Mr. Karl Howell']26.0age.mean()29.69911764705882df_titanic_raw[age < 2]| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | |||||||||||
| Caldwell, Master. Alden Gates | 79 | 1 | 2 | male | 0.83 | 0 | 2 | 248738 | 29.0000 | NaN | S |
| Panula, Master. Eino Viljami | 165 | 0 | 3 | male | 1.00 | 4 | 1 | 3101295 | 39.6875 | NaN | S |
| Johnson, Miss. Eleanor Ileen | 173 | 1 | 3 | female | 1.00 | 1 | 1 | 347742 | 11.1333 | NaN | S |
| Becker, Master. Richard F | 184 | 1 | 2 | male | 1.00 | 2 | 1 | 230136 | 39.0000 | F4 | S |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Dean, Master. Bertram Vere | 789 | 1 | 3 | male | 1.00 | 1 | 2 | C.A. 2315 | 20.5750 | NaN | S |
| Thomas, Master. Assad Alexander | 804 | 1 | 3 | male | 0.42 | 0 | 1 | 2625 | 8.5167 | NaN | C |
| Mallet, Master. Andre | 828 | 1 | 2 | male | 1.00 | 0 | 2 | S.C./PARIS 2079 | 37.0042 | NaN | C |
| Richards, Master. George Sibley | 832 | 1 | 2 | male | 0.83 | 1 | 1 | 29106 | 18.7500 | NaN | S |
14 rows × 11 columns
# You can come back to the original indexing
df_titanic_raw = df_titanic_raw.reset_index()Counting values for categorical variables
df_titanic_raw['Embarked'].value_counts(normalize=False, sort=True,
ascending=False)Embarked
S 644
C 168
Q 77
Name: count, dtype: int64pd.options.display.max_rows = 70
df_titanic[df_titanic['Embarked'] == 'C']
dir(numpy)| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 52 | 53 | 1 | 1 | Harper, Mrs. Henry Sleeper (Myna Haxtun) | female | 49.0 | 1 | 0 | PC 17572 | 76.7292 | D33 | C |
| 54 | 55 | 0 | 1 | Ostby, Mr. Engelhart Cornelius | male | 65.0 | 0 | 1 | 113509 | 61.9792 | B30 | C |
| 96 | 97 | 0 | 1 | Goldschmidt, Mr. George B | male | 71.0 | 0 | 0 | PC 17754 | 34.6542 | A5 | C |
| 97 | 98 | 1 | 1 | Greenfield, Mr. William Bertram | male | 23.0 | 0 | 1 | PC 17759 | 63.3583 | D10 D12 | C |
| 118 | 119 | 0 | 1 | Baxter, Mr. Quigg Edmond | male | 24.0 | 0 | 1 | PC 17558 | 247.5208 | B58 B60 | C |
| 139 | 140 | 0 | 1 | Giglio, Mr. Victor | male | 24.0 | 0 | 0 | PC 17593 | 79.2000 | B86 | C |
| 174 | 175 | 0 | 1 | Smith, Mr. James Clinch | male | 56.0 | 0 | 0 | 17764 | 30.6958 | A7 | C |
| 177 | 178 | 0 | 1 | Isham, Miss. Ann Elizabeth | female | 50.0 | 0 | 0 | PC 17595 | 28.7125 | C49 | C |
| 194 | 195 | 1 | 1 | Brown, Mrs. James Joseph (Margaret Tobin) | female | 44.0 | 0 | 0 | PC 17610 | 27.7208 | B4 | C |
| 195 | 196 | 1 | 1 | Lurette, Miss. Elise | female | 58.0 | 0 | 0 | PC 17569 | 146.5208 | B80 | C |
| 209 | 210 | 1 | 1 | Blank, Mr. Henry | male | 40.0 | 0 | 0 | 112277 | 31.0000 | A31 | C |
| 215 | 216 | 1 | 1 | Newell, Miss. Madeleine | female | 31.0 | 1 | 0 | 35273 | 113.2750 | D36 | C |
| 218 | 219 | 1 | 1 | Bazzani, Miss. Albina | female | 32.0 | 0 | 0 | 11813 | 76.2917 | D15 | C |
| 273 | 274 | 0 | 1 | Natsch, Mr. Charles H | male | 37.0 | 0 | 1 | PC 17596 | 29.7000 | C118 | C |
| 291 | 292 | 1 | 1 | Bishop, Mrs. Dickinson H (Helen Walton) | female | 19.0 | 1 | 0 | 11967 | 91.0792 | B49 | C |
| 292 | 293 | 0 | 2 | Levy, Mr. Rene Jacques | male | 36.0 | 0 | 0 | SC/Paris 2163 | 12.8750 | D | C |
| 299 | 300 | 1 | 1 | Baxter, Mrs. James (Helene DeLaudeniere Chaput) | female | 50.0 | 0 | 1 | PC 17558 | 247.5208 | B58 B60 | C |
| 307 | 308 | 1 | 1 | Penasco y Castellana, Mrs. Victor de Satode (M... | female | 17.0 | 1 | 0 | PC 17758 | 108.9000 | C65 | C |
| 309 | 310 | 1 | 1 | Francatelli, Miss. Laura Mabel | female | 30.0 | 0 | 0 | PC 17485 | 56.9292 | E36 | C |
| 310 | 311 | 1 | 1 | Hays, Miss. Margaret Bechstein | female | 24.0 | 0 | 0 | 11767 | 83.1583 | C54 | C |
| 311 | 312 | 1 | 1 | Ryerson, Miss. Emily Borie | female | 18.0 | 2 | 2 | PC 17608 | 262.3750 | B57 B59 B63 B66 | C |
| 319 | 320 | 1 | 1 | Spedden, Mrs. Frederic Oakley (Margaretta Corn... | female | 40.0 | 1 | 1 | 16966 | 134.5000 | E34 | C |
| 325 | 326 | 1 | 1 | Young, Miss. Marie Grice | female | 36.0 | 0 | 0 | PC 17760 | 135.6333 | C32 | C |
| 329 | 330 | 1 | 1 | Hippach, Miss. Jean Gertrude | female | 16.0 | 0 | 1 | 111361 | 57.9792 | B18 | C |
| 337 | 338 | 1 | 1 | Burns, Miss. Elizabeth Margaret | female | 41.0 | 0 | 0 | 16966 | 134.5000 | E40 | C |
| 366 | 367 | 1 | 1 | Warren, Mrs. Frank Manley (Anna Sophia Atkinson) | female | 60.0 | 1 | 0 | 110813 | 75.2500 | D37 | C |
| 369 | 370 | 1 | 1 | Aubart, Mme. Leontine Pauline | female | 24.0 | 0 | 0 | PC 17477 | 69.3000 | B35 | C |
| 370 | 371 | 1 | 1 | Harder, Mr. George Achilles | male | 25.0 | 1 | 0 | 11765 | 55.4417 | E50 | C |
| 377 | 378 | 0 | 1 | Widener, Mr. Harry Elkins | male | 27.0 | 0 | 2 | 113503 | 211.5000 | C82 | C |
| 393 | 394 | 1 | 1 | Newell, Miss. Marjorie | female | 23.0 | 1 | 0 | 35273 | 113.2750 | D36 | C |
| 452 | 453 | 0 | 1 | Foreman, Mr. Benjamin Laventall | male | 30.0 | 0 | 0 | 113051 | 27.7500 | C111 | C |
| 453 | 454 | 1 | 1 | Goldenberg, Mr. Samuel L | male | 49.0 | 1 | 0 | 17453 | 89.1042 | C92 | C |
| 473 | 474 | 1 | 2 | Jerwan, Mrs. Amin S (Marie Marthe Thuillard) | female | 23.0 | 0 | 0 | SC/AH Basle 541 | 13.7917 | D | C |
| 484 | 485 | 1 | 1 | Bishop, Mr. Dickinson H | male | 25.0 | 1 | 0 | 11967 | 91.0792 | B49 | C |
| 487 | 488 | 0 | 1 | Kent, Mr. Edward Austin | male | 58.0 | 0 | 0 | 11771 | 29.7000 | B37 | C |
| 496 | 497 | 1 | 1 | Eustis, Miss. Elizabeth Mussey | female | 54.0 | 1 | 0 | 36947 | 78.2667 | D20 | C |
| 505 | 506 | 0 | 1 | Penasco y Castellana, Mr. Victor de Satode | male | 18.0 | 1 | 0 | PC 17758 | 108.9000 | C65 | C |
| 523 | 524 | 1 | 1 | Hippach, Mrs. Louis Albert (Ida Sophia Fischer) | female | 44.0 | 0 | 1 | 111361 | 57.9792 | B18 | C |
| 539 | 540 | 1 | 1 | Frolicher, Miss. Hedwig Margaritha | female | 22.0 | 0 | 2 | 13568 | 49.5000 | B39 | C |
| 544 | 545 | 0 | 1 | Douglas, Mr. Walter Donald | male | 50.0 | 1 | 0 | PC 17761 | 106.4250 | C86 | C |
| 550 | 551 | 1 | 1 | Thayer, Mr. John Borland Jr | male | 17.0 | 0 | 2 | 17421 | 110.8833 | C70 | C |
| 556 | 557 | 1 | 1 | Duff Gordon, Lady. (Lucille Christiana Sutherl... | female | 48.0 | 1 | 0 | 11755 | 39.6000 | A16 | C |
| 581 | 582 | 1 | 1 | Thayer, Mrs. John Borland (Marian Longstreth M... | female | 39.0 | 1 | 1 | 17421 | 110.8833 | C68 | C |
| 583 | 584 | 0 | 1 | Ross, Mr. John Hugo | male | 36.0 | 0 | 0 | 13049 | 40.1250 | A10 | C |
| 587 | 588 | 1 | 1 | Frolicher-Stehli, Mr. Maxmillian | male | 60.0 | 1 | 1 | 13567 | 79.2000 | B41 | C |
| 591 | 592 | 1 | 1 | Stephenson, Mrs. Walter Bertram (Martha Eustis) | female | 52.0 | 1 | 0 | 36947 | 78.2667 | D20 | C |
| 599 | 600 | 1 | 1 | Duff Gordon, Sir. Cosmo Edmund ("Mr Morgan") | male | 49.0 | 1 | 0 | PC 17485 | 56.9292 | A20 | C |
| 632 | 633 | 1 | 1 | Stahelin-Maeglin, Dr. Max | male | 32.0 | 0 | 0 | 13214 | 30.5000 | B50 | C |
| 641 | 642 | 1 | 1 | Sagesser, Mlle. Emma | female | 24.0 | 0 | 0 | PC 17477 | 69.3000 | B35 | C |
| 645 | 646 | 1 | 1 | Harper, Mr. Henry Sleeper | male | 48.0 | 1 | 0 | PC 17572 | 76.7292 | D33 | C |
| 647 | 648 | 1 | 1 | Simonius-Blumer, Col. Oberst Alfons | male | 56.0 | 0 | 0 | 13213 | 35.5000 | A26 | C |
| 659 | 660 | 0 | 1 | Newell, Mr. Arthur Webster | male | 58.0 | 0 | 2 | 35273 | 113.2750 | D48 | C |
| 679 | 680 | 1 | 1 | Cardeza, Mr. Thomas Drake Martinez | male | 36.0 | 0 | 1 | PC 17755 | 512.3292 | B51 B53 B55 | C |
| 681 | 682 | 1 | 1 | Hassab, Mr. Hammad | male | 27.0 | 0 | 0 | PC 17572 | 76.7292 | D49 | C |
| 698 | 699 | 0 | 1 | Thayer, Mr. John Borland | male | 49.0 | 1 | 1 | 17421 | 110.8833 | C68 | C |
| 700 | 701 | 1 | 1 | Astor, Mrs. John Jacob (Madeleine Talmadge Force) | female | 18.0 | 1 | 0 | PC 17757 | 227.5250 | C62 C64 | C |
| 710 | 711 | 1 | 1 | Mayne, Mlle. Berthe Antonine ("Mrs de Villiers") | female | 24.0 | 0 | 0 | PC 17482 | 49.5042 | C90 | C |
| 716 | 717 | 1 | 1 | Endres, Miss. Caroline Louise | female | 38.0 | 0 | 0 | PC 17757 | 227.5250 | C45 | C |
| 737 | 738 | 1 | 1 | Lesurer, Mr. Gustave J | male | 35.0 | 0 | 0 | PC 17755 | 512.3292 | B101 | C |
| 742 | 743 | 1 | 1 | Ryerson, Miss. Susan Parker "Suzette" | female | 21.0 | 2 | 2 | PC 17608 | 262.3750 | B57 B59 B63 B66 | C |
| 789 | 790 | 0 | 1 | Guggenheim, Mr. Benjamin | male | 46.0 | 0 | 0 | PC 17593 | 79.2000 | B82 B84 | C |
| 835 | 836 | 1 | 1 | Compton, Miss. Sara Rebecca | female | 39.0 | 1 | 1 | PC 17756 | 83.1583 | E49 | C |
| 879 | 880 | 1 | 1 | Potter, Mrs. Thomas Jr (Lily Alexenia Wilson) | female | 56.0 | 0 | 1 | 11767 | 83.1583 | C50 | C |
| 889 | 890 | 1 | 1 | Behr, Mr. Karl Howell | male | 26.0 | 0 | 0 | 111369 | 30.0000 | C148 | C |
Comments: not all passengers from Cherbourg are Gallic (🇫🇷: gaulois) …
What is the survival rate for raw data?
df_titanic_raw['Survived'].mean()0.3838383838383838What is the survival rate for data after removing missing values?
df_titanic['Survived'].mean()0.6721311475409836See also the command:
df_titanic.groupby(['Sex'])[['Survived', 'Age', 'Fare']].mean()| Survived | Age | Fare | |
|---|---|---|---|
| Sex | |||
| female | 0.931818 | 32.676136 | 89.000900 |
| male | 0.431579 | 38.451789 | 69.124343 |
Conclusion: Be careful when you remove some missing values, the missingness might be informative!
Data import/export
The Pandas library supports many formats:
- CSV, text
- SQL database
- Excel
- HDF5
- JSON
- HTML
- pickle
- sas, stata
- …
Data Exploration
You can explore the data starting from the top or the bottom of the file:
Access values by line (iloc) or columns (loc)
Theloc and iloc functions select indices: - the loc function selects rows using row labels whereas - the iloc function selects rows using their integer positions.
iloc
df_titanic_raw.iloc[0:2]| Name | PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Braund, Mr. Owen Harris | 1 | 0 | 3 | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | 2 | 1 | 1 | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
loc
# with naming indexing
df_titanic_raw = df_titanic_raw.set_index('Name') # You can only do it once !!
df_titanic_raw.loc['Bonnell, Miss. Elizabeth', 'Fare']26.55df_titanic_raw.loc['Bonnell, Miss. Elizabeth']PassengerId 12
Survived 1
Pclass 1
Sex female
Age 58.0
SibSp 0
Parch 0
Ticket 113783
Fare 26.55
Cabin C103
Embarked S
Name: Bonnell, Miss. Elizabeth, dtype: objectdf_titanic_raw.loc['Bonnell, Miss. Elizabeth', 'Survived']
df_titanic_raw.loc['Bonnell, Miss. Elizabeth', 'Survived'] = 0df_titanic_raw.loc['Bonnell, Miss. Elizabeth']PassengerId 12
Survived 0
Pclass 1
Sex female
Age 58.0
SibSp 0
Parch 0
Ticket 113783
Fare 26.55
Cabin C103
Embarked S
Name: Bonnell, Miss. Elizabeth, dtype: object# set back the original value
df_titanic_raw.loc['Bonnell, Miss. Elizabeth', 'Survived'] = 1
df_titanic_raw = df_titanic_raw.reset_index() # Back to original indexingCreate binned values
bins=np.arange(0, 100, 10)
current_palette = sns.color_palette()
df_test = pd.DataFrame({ 'Age': pd.cut(df_titanic['Age'], bins=bins, right=False)})
ax = sns.countplot(data=df_test, x='Age', color=current_palette[0])
ax.tick_params(axis='x', labelrotation=90)
plt.title("Binning age per decades")
plt.show()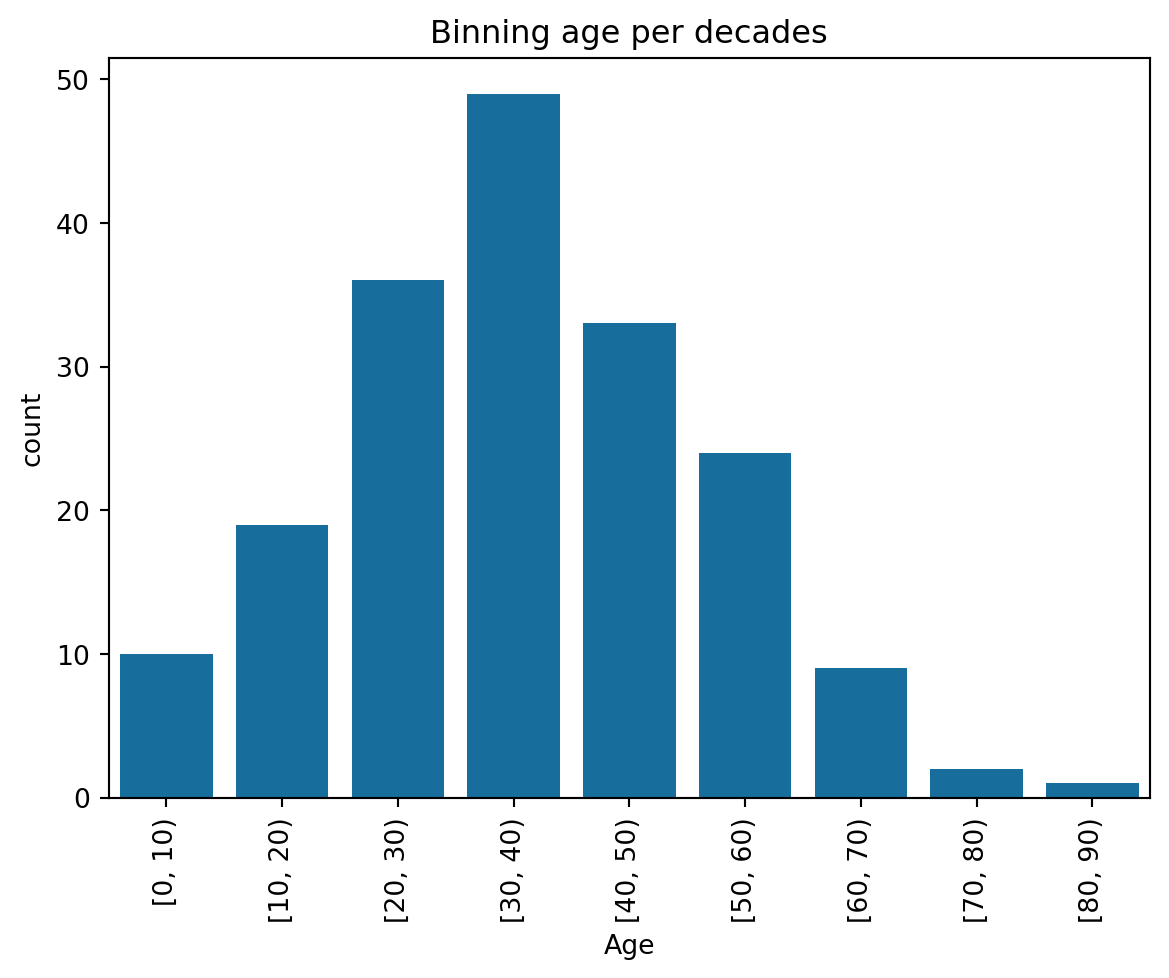
References
Other interactive tools for data visualization include Altair, Bokeh, etc. See comparisons by Aaron Geller: link
How to choose your chart by A ndrew V. Abela.